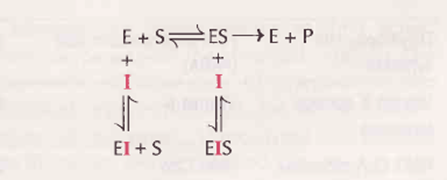Enzyme Induction and Enzyme Inhibition

Enzyme Induction and Enzyme Inhibition
Objective
At the end of this lecture, student will be able to
• Explain enzyme Induction
• Explain enzyme inhibition
Enzyme Inhibition
• Enzyme inhibitor is defined as substance which binds with the enzyme and brings about a decrease in catalyrtc activity of that enzyme
• The inhibitor may be organic or inorganic in nature
• There are three broad categories of enzyme inhibition
1. Reversible inhibition
2. Irreversible inhibition
3. Allosteric inhibition
1. Reversible inhibition
• The inhibiior binds non-covalently with enzyme and the enzyme inhibition can be reversed if the inhibitor is removed
• Further sub-divided into
l. Competitive inhibition
ll. Non-competitive inhibition
l. Competitive inhibition: The inhibitor (l) which closely resembles the real substrate (S) is regarded as a substrate analogue
• The inhibitor competes with substrate and binds at the active site of the enzyme but does not undergo any catalysis. As long as the competitive inhibitor
• As long as the competitive inhibitor holds the active site, the enzyme is not available for the substrate to bind
• The relative concentration of the substrate and inhibitor and their respective affinity with the enzyme determines the degree of competitive inhibition
• The inhibition could be overcome by a high substrate concentration
• ln competitive inhibition, the KM-value increases whereas Vmax remains unchanged
• The enzyme succinate dehydrogenase (SDH) is a classical example of competitive inhibition with succinic acid as its substrate.
• Methanol is toxic to the body when it is converted to formaldehyde by the enzyme alcohol dehydrogenase (ADH). Ethanol can compete with methanol for ADH. Thus, ethanol can be used in the treatment of methanol poisoning
Some examples of competitive inhibitors are
ll. Non-competitive inhibition:
• The inhibitor binds at a site other than the active site on the enzyme surface
• This binding impairs the enzyme function
• The inhibitor has no structural resemblance with the substrate, However exists a strong affinity for the inhibitor to bind at the second site
• Inhibitor does not interfere with the enzyme-substrate binding, but the catalysis is prevented, due to distortion in the enzyme conformation
• The inhibitor generally binds with the enzyme as well as the ES complex
• For non-competitive inhibition, the KM value is unchanged while Vmax is lowered
• E.g. Heavy metal ions (Ag+, Pb2+, Hg2+ etc) can non-competitively inhibit the enzymes by binding with cysteinyl sulfhydryl groups
• The general reaction for Hg2+ is shown below
2. irreversible inhibition
• The inhibitors bind covalently with the enzymes and inactivate them, which is irreversible
• These inhibitors are usually toxic poisonous substances
• Iodoacetate is an irreversible inhibitor of the enzymes like papain and glyceraldehyde 3-phosphate dehydrogenase, here Iodoacetate combines with sulfhydryl (-SH) groups at the active site of these enzymes and makes them inactive
• Di-isopropyl fluorophosphate (DFP) is a nerve gas developed by the Germans during Second World War. DFP irreversibly binds with enzymes containing serine at the active site, e.g. Serine proteases, acetylcholine esterase.
• Many organophosphorus insecticides like melathion are toxic to animals (including man) as they block the activity of acetylcholine esterase (essential for nerve conduction), resulting in paralysis of vital body functions
• Disulfiram used in the treatment of alcoholism, irreversibly inhibits the enzyme aldehyde dehydrogenase
• Penicillin antibiotics act as irreversible inhibitors of serine-containing enzymes and block the bacterial cell wall synthesis
• lrreversible inhibitors are frequently used to identify amino acid residues at the active site of the enzymes and also to understand the mechanism of enzyme action
3. Allosteric inhibition
• The details of this type of inhibition are given under allosteric regulation as a part of the regulation of enzyme activity in the living system
• Some enzymes are called allosteric enzymes, they exist in alternate higher order structures
• Binding of ligand can either enhance the activity of the enzyme or inhibit it
• The allosteric sites are unique places on the enzyme molecule
• Allosteric enzymes possess a site distinct and physically separate from the substrate binding site and is called as allosteric site.
• Allosteric modulators bind to allosteric site by reversible, non-covalent interactions.
Allosteric effectors
• Certain substances referred to as allosteric modulators (effectors or modifiers) bind at the allosteric site and regulate the enzyme activity. The enzyme activity is increased when a positive (+) allosteric effector binds at the allosteric site known as activator site.
• On the other hand, a negative (-) allosteric effector binds at the allosteric site called inhibitor site and inhibits the enzyme activity
The allosteric site at which the positive modulator binds is referred to as an activator site, the negative modulator binds at an inhibitory site.
Classes of allosteric enzymes
• Enzymes that are regulated by allosteric mechanism are referred to as allosteric enzymes.
• They are divided into two classes based on the influence of allosteric effector on Km, and Vmax:
K-class of allosteric enzymes
V-class of allosteric enzymes
Enzyme Induction
Enzyme specificity
• Enzymes are highly specific in their action when compared with the chemical catalysts
• 3 types of enzyme specificity are well-recognised
1. Stereospecificity
2. Reaction specificity
3. Substrate specificity
1. Stereospecificity or optical specificity:
• Stereoisomers are the comoounds which have the same molecular formula, but differ in their structural configuration
• The enzymes act only on one isomer and therefore, exhibit stereospecificity
e.g. L-amino acid oxidase and D-amino acid oxidase act on L- and D-amino acids respectively
Hexokinase acts on D-hexoses; Glucokinase on D-glucose
• The class of enzymes belonging to isomerases do not exhihit stereospecificity since they are specializedin the interconversion of isomers
2. Reaction specificity:
• The same substrate can undergo different types of reactions, each catalysed by a separate enzyme and this is referred to as reaction specificity
e.g. Amino acid undergo transamination, oxidative deamination, decarboxylation, racemization etc.
3. Substrate specificity:
• The substrate specificity varies from enzyme to enzyme
• lt may be either absolute, relative or broad
Absolute substrate specificity:
• Certain enzymes act only on one substrate e.g. glucokinase acts on glucose to give glucose-6-phosphate, urease cleaves urea to ammonia and carbon dioxide
Relative substrate specificity:
• Some enzymes act on structurally related substances, may be dependent on the specific group or a bond present
• Gorup specificity
• e.g. Trypsin hydrolyses peptide linkage involving arginine or lysine Chymotrypsin cleaves peptide bonds attached to aromatic amino acids (phenylalanine, tyrosine and tryptophan)
Bond specificity:
Examples of bond specificity specificity glycosidases acting on glycosidic bonds of carbohydrates, lipases cleaving ester bonds of lipids etc
Broad specificity:
• Some enzymes act on closely related substrates which is commonly known as broad substrate specificity
• e.g. hexokinase acts on glucose, fructose/mannose and glucosamine and not on galactose
Mechanism of enzyme action
• Catalysis is the prime function of enzymes, for any chemical reaction to occur, the reactants have to be in an activated state
• The energy required by the reactants to undergo the reaction is known as activation energy
• The reactants when heated attain the activation energy & enzyme in the biological system reduces the activation energy and this causes the reaction to proceed at a lower temperature
• Enzymes do not alter the equilibrium constants, they only enhance the velocity of the reaction
• The enzymes reduce the activation energy of the reactants in biological systems to undergo at body temperature (below 40oC)
• Enzyme-substrate complex formation:
• Substrate (S) must combine with the enzyme (E) at the active site to form enzyme-substrate complex (ES) which ultimately results in the product formation (P)
E + S↔ES + E + P
• A few theories have been put forth to explain mechanism of enzyme-substrate complex formation
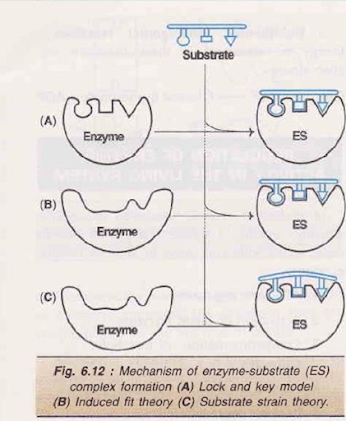
Lock and key model or Fischer’s template theory
• By German biochemist Emil Fischer
• According to this model, the structure or conformation of the enzyme is rigid
• The substrate fits to the binding site just as a key fits into the proper lock
• Thus the active site of an enzyme is a rigid and pre-shaped template where only a specific substrate can bind
• This model does not give any scope for the flexible nature of enzymes, hence the model totally fails to explain many facts of enzymatic reactions
Induced fit theory or Koshland’s model
• By Koshland, in 1958, proposed a more acceptable model for ES complex formation
• According to this model, the active site is not rigid and pre-shaped
• The interaction of the substrate with the enzyme induces a fit or a conformation change in the enzyme, resulting in the formation of a strong substrate binding site
• Further, due to induced fit, the appropriate amino acids of the enzyme are repositioned to form the actives and bring about the catalysis
Substrate strain theory
• In this model, the substrate is strained due conformation change in the enzyme to the induced
• When a substrate binds to the preformed active induces a strain to the substrate site, the enzyme
• The strained substrate leads to the formation of product
• Therefore, combination of the induced fit model with the substrates train is considered to be operative in the enzymatic action
Summary
• Enzyme inhibitor is defined as substance which binds with the enzyme and brings about a decrease in catalyrtc activity of that enzyme
• Enzyme inhibition are of their type’s Reversible inhibition, Irreversible inhibition & Allosteric inhibition
• Enzymes are highly specific in their action when compared with the chemical catalysts
• 3 types of enzyme specificity are well-recognised, Stereospecificity, Reaction specificity & Substrate specificity
• Lock and key model, Induced fit theory & Substrate strain theory have been put forth to explain mechanism of enzyme-substrate complex formation
FAQs
- What is the primary difference between enzyme induction and enzyme inhibition?
- Enzyme induction stimulates the synthesis of enzymes, adapting to changing conditions, while enzyme inhibition reduces or halts enzyme activity, often for therapeutic purposes.
- How do genetic factors influence enzyme regulation?
- Genetic variations can lead to differences in enzyme activity, impacting an individual’s response to drugs and environmental factors.
- Can lifestyle choices affect enzyme activity?
- Yes, diet and lifestyle choices can either induce or inhibit enzymes, influencing overall health and well-being.
- What role does enzyme modulation play in drug development?
- Enzyme modulation is crucial in drug design, allowing researchers to target specific pathways for therapeutic effects while minimizing side effects.
- How does personalized medicine utilize knowledge of enzyme regulation?
- Personalized medicine tailors medical interventions based on individual enzyme profiles, optimizing treatment outcomes for patients.
Also, Visit:
B. Pharma Notes | B. Pharma Notes | Study material Bachelor of Pharmacy pdf