Prokaryotic chromosome
structure and Chromatin
Objectives
At the end of this lecture,
student will be able to describe the following
• Chromosome structure of prokaryotes in general
• Chromosome structure of E. coli
• Chromatin structure
Content
• Prokaryotic Chromosome structure
• Chromatin structure
Prokaryotic
chromosome structure
• Prokaryotic genome vary in size
• 0.16 Mb of endosymbiont (Carsonella ruddi) to 13 Mb of
soil bacterium (Sorangium cellulosum)
• Chromosome with single circular DNA molecule
• 4.6 Mb of E. coli chromosome
• DNA is packed in a region of cell
• Unbounded nucleiod
• Has very high DNA concentration, along with certain
proteins
• Has single or multiple copies of additional small circular
DNA molecule – Plasmids
• Vibrio cholerae –
1 Mb circle of DNA + 3 Mb chromosome (Second chromosome)
• Agrobacterium
tumefaciens have 2.9 Mb circle + 2.1 Mb linear DNA +0.5 Mb + 0.3Mb circular
plasmid
• Ti plasmid used as cloning vector
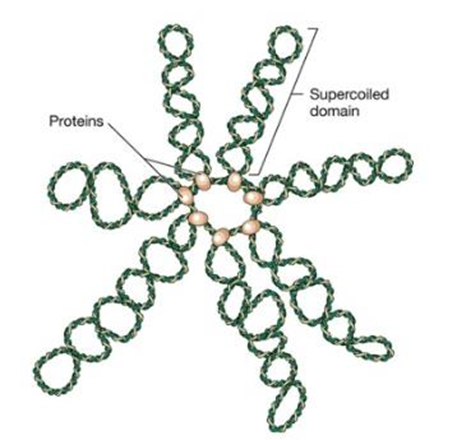
DNA domains
• Consists of 50-100 domains or loops
• Ends are constrained by binding to structure made of
proteins attached to cell membrane
• Loops – 50-100 Kb in size
• Loops may spool through the sites of polymerase or other
enzymic action at the base of loops
Chromosome
structure
Super
coiling of the genome
• E. Coli chromosome is negatively supercoiled
• Individual domains may be supercoiled independently
• Some domains may not be supercoiled
• DNA has become broken in one strand
• Other domains clearly do contain supercoils
• Attachment of the DNA to the protein
• Membrane scaffold act as a barrier to rotation of the DNA
• Domains may be topologically independent
DNA binding
protein
• Abundant protein -HU, a small basic (positively charged) protein
• Binds DNA nonspecifically by the wrapping of the DNA around
the protein
• H-NS (protein H1)- a monomeric neutral protein
• binds DNA nonspecifically in terms of sequence
• Also known as histone-like proteins
• Compacting the DNA – essential for the packaging of the
DNA into the nucleoid
• Stabilizes and constrain the supercoiling of the
chromosome
• Half of this is constrained as permanent wrapping of DNA around
proteins such as HU
• About half the supercoiling is unconstrained
• RNA polymerase and mRNA molecules, site-specific DNA- binding
proteins such as integration host factor (IHF), a homolog of HU, which binds to
specific DNA sequences and bends DNA through 140°
Chromatin
structure (Chromatin)
• Highly organized complex of DNA, nucleoprotein complex
• Makes up eukaryotic chromosome
• Serves to package and organize the chromosomal DNA
• Alters its level of packaging at different stages of cell
cycle
Histones
• Major protein components of chromatin
• Most of the protein in eukaryotic chromatin consists of
histones
• Five families, or classes of histones-H2A, H2B, H3 and H4:
core histones
• Core histones are small proteins, with masses between 10
and 20 kDa
• H1: little larger at around 23 kda
• All histone proteins have a large positive charge
• 20 – 30% of their sequences consist of the basic amino
acids, lysine and arginine
• Histones will bind very strongly to the negatively charged
DNA in forming chromatin
Nucleosome
• Basic unit of chromatin
• Composed of approximately 146 base pairs of DNA wrapped in
1.8 helical turns around an eight-unit structure-histone protein octamer
• Octamer consists of two copies each of the histones h2a,
h2b, H3, and H4
• Space in between individual nucleosomes is referred to as linker
DNA
• Linker DNA interacts with the linker histone – H1
• About 142 hydrogen bonds are formed between DNA and the histone
core
• Wrapping of DNA is responsible for negative supercoiling
in eukaryotic DNA
Role of H1
• A single molecule of H1 stabilises the DNA at the point at
which it enter and leaves the nucleosome core
• Organises DNA between nucleosome
• Nucleosome+H1 – Chromatosome
• In some cases H1 is replace by H5
• Binds more tightly
• Associated with DNA which is inactive in transcription
Linker
DNA
• Linker DNA between nucleosome varies betweem > 10 &
< 100 bp
• Normally around 55bp
• Nucleosomal repeat unit is around 200 bp
• Comprises of globular protein connected by thin strands of
DNA
The 30-nm
Fiber
• Large structure of organised chromatin
• Also known as solenoid
• Consists of left handed helix of nucleosome
• Approximately 6 nucleosome per helical structure
• Most chromosomal DNA in vivo is packaged into 30-nm fibre
Nucleosome
and associated structures
Higher
order structure
• Organization of chromatin at the highest level
• Chromosomal DNA is organized into loops of up to 100 kb
• In the form of 30-nm fiber
• Constrained by a protein scaffold, the nuclear matrix
• Overall structure resembles organizational domain of
prokaryotic DNA
Summary
• Prokaryotic chromosome consists of a single circular DNA
molecule
• It consists of 50-100 domains or loops of DNA domains that
may be supercoiled
• DNA binding protein binds the DNA nonspecifically by the
wrapping it around the protein
• Chromatin is a highly organized complex of DNA,
nucleoprotein complex
• Consists of histones, linker DNA, 30-nm fibre and
nucleosome
• The whole of chromatin is organized into higher order structure
similar to that of prokaryotic DNA